Genomic variant analysis platform powered by the revolutionary Evo2 DNA language model. Enables researchers and clinicians to predict genetic variant pathogenicity with unprecedented accuracy using 1M+ base pair context modeling.
- Repositoryhttps://github.com/insertfahim/variant-analysis-evo2
- PlatformWeb (Next.js) / GPU Cloud Computing (Modal)
- Frontend StackNext.js 15, TypeScript, Tailwind CSS, Radix UI
- Backend StackPython, FastAPI, Modal Serverless, Evo2 AI Model
- AI/MLEvo2 DNA Language Model, StripedHyena Architecture, H100 GPUs
- Data SourcesNCBI Gene API, ClinVar, UCSC Genome Browser

Key Features
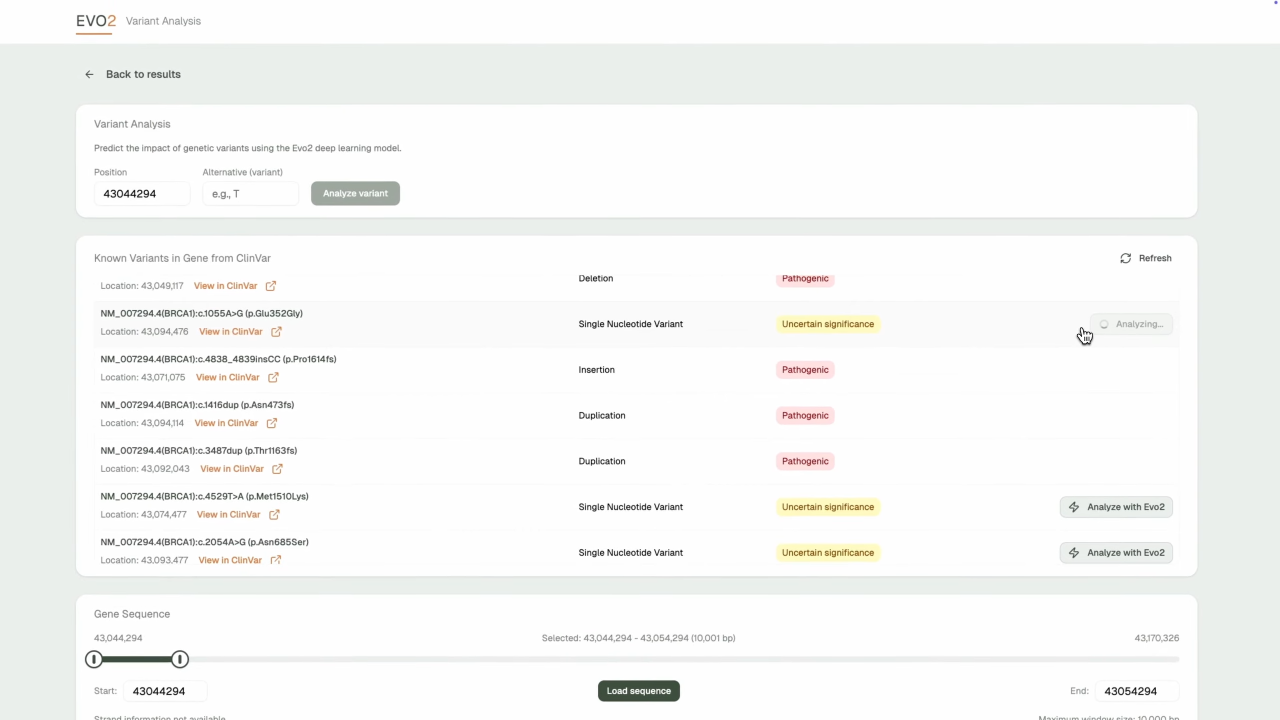
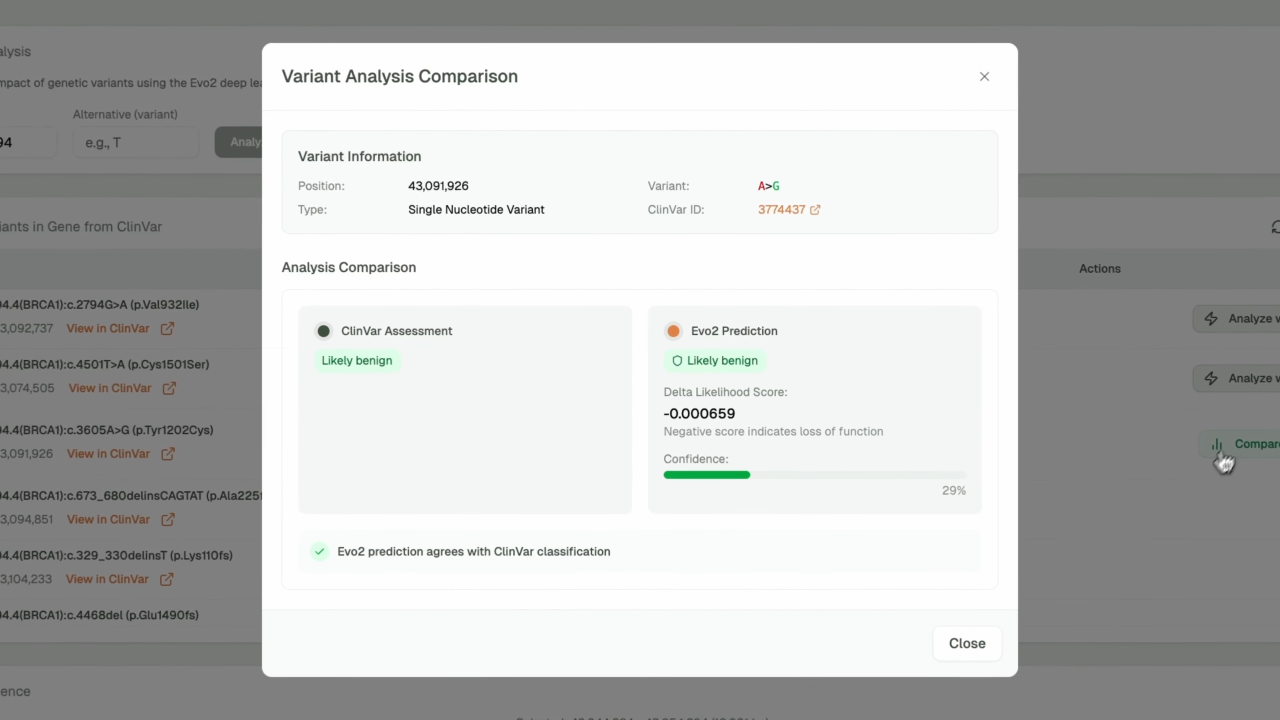
- AI-Powered Variant Prediction: Leverages Evo2's deep learning model for accurate pathogenicity assessment with confidence scoring
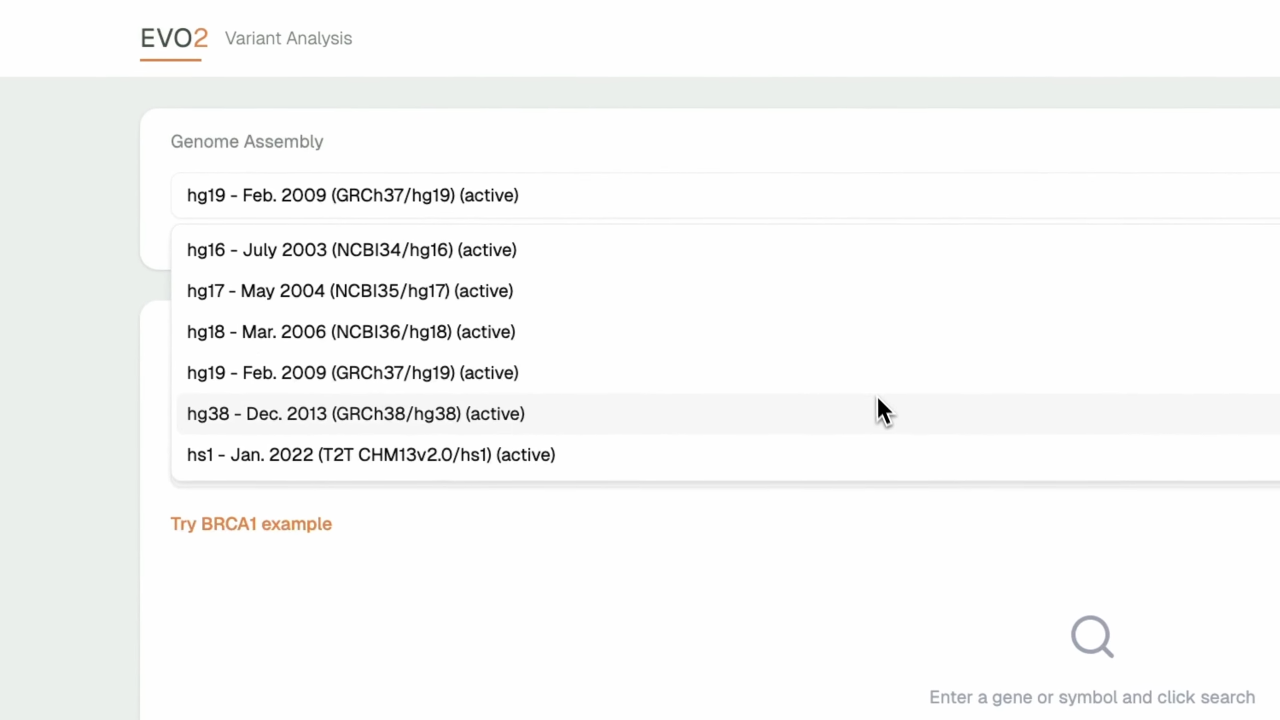
- Multi-Genome Support: Compatible with hg19, hg38, and other genome assemblies for comprehensive analysis
- Real-Time Processing: Instant analysis with GPU acceleration via Modal serverless infrastructure
- Interactive Visualization: Color-coded nucleotide display with positional mapping and sequence viewer
- Clinical Integration: ClinVar database integration with known variant classifications and clinical significance
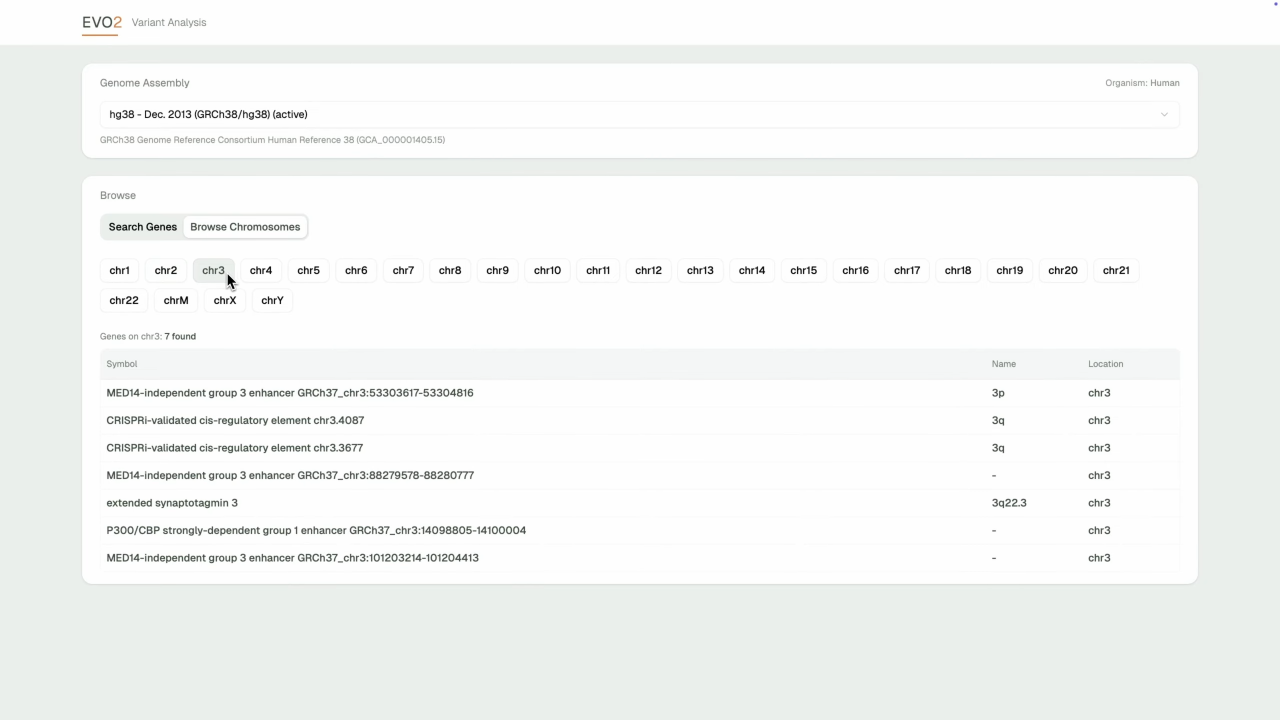
- Intelligent Search: NCBI-powered gene discovery with autocomplete and chromosome browsing capabilities
Technical Architecture
The platform features a full-stack architecture with a modern Next.js frontend and a high-performance Python backend. The AI inference is powered by Modal's serverless GPU infrastructure, providing auto-scaling compute resources for the Evo2 model processing.
Innovation Highlights
- First platform to implement Evo2's 1M+ base pair context modeling for genomic analysis
- Revolutionary StripedHyena hybrid attention-convolution architecture integration
- Serverless GPU computing with pay-per-use H100 GPU acceleration
- Real-time genomic sequence analysis with clinical-grade accuracy
- Comprehensive API integration with major genomic databases
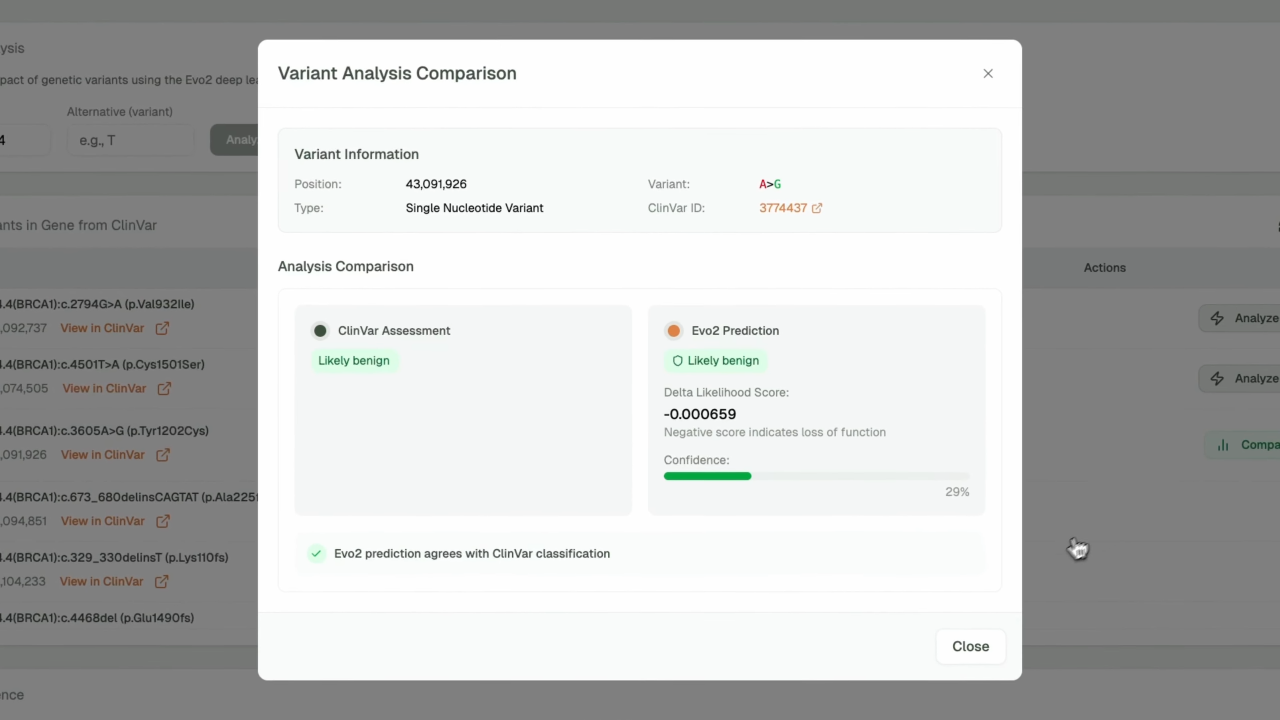
This platform represents a significant advancement in making cutting-edge genomic AI accessible to researchers and clinicians worldwide, bridging the gap between state-of-the-art machine learning research and practical clinical applications.